Introduction
Overview
The Deep DNAshape websever can be used for quickly checking 14 DNA shape features and 13 shape fluctuations for any sequences (one or many), using query tables pre-computed from Deep DNAshape models. Compared to the deep learning model, using this webserver doesn't require downloading and installing tensorflow and running any codes. This web-server has pre-computed enough data from the Deep DNAshape model for instantaneously prediciton.
The Deep DNAshape webserver is an updated version of previous popular DNAshape webserver used to predict DNA shape features given one or more sequences. The previous webserver can still be found at the pentamer-based DNAShape Webserver.
DNA shape features are derived DNA structural parameters for quantitatively analysis of DNA structures. For detail informaiton about DNA shape features, please take a look at this paper. DNA shape fluctuation measures how easy the static DNA shape can be spontaneuosly altered, which indicates DNA flexibility in DNA shape level. The web server is able to predict 14 DNA shape features, including 2 DNA groove features, 6 intra-base-pair features and 6 inter-base-pair features; 13 DNA shape fluctuation features, excluding EP-FL.
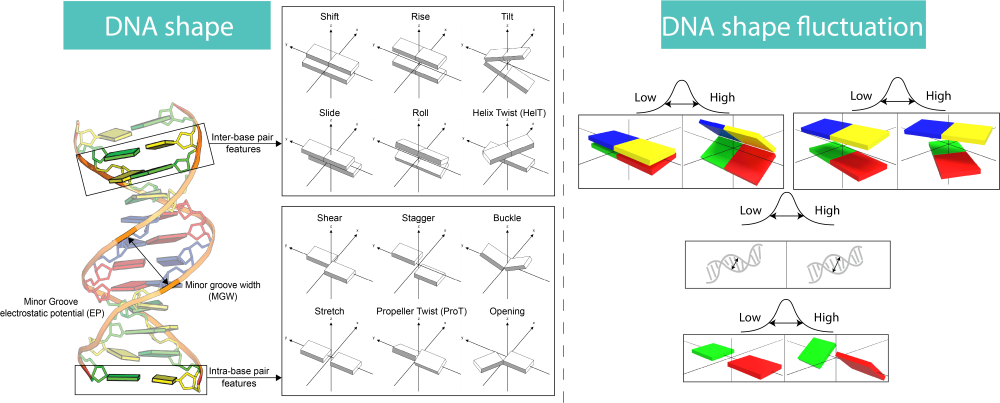
Getting Started
Single sequence
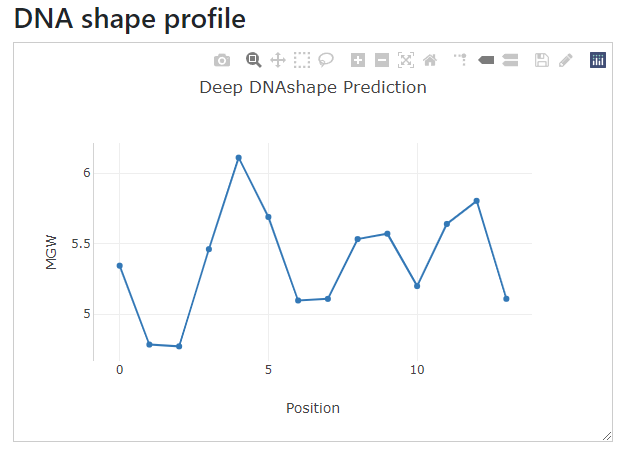
In the left text input box, input:
ACGTCACGTGGTAG

The right side will automatically have the DNA shape profile drawn.
Now, we can edit the sequence by changing any position to another character from A, C, G, T and N. The right panel will reflect the change simultaneously.
Multiple sequences
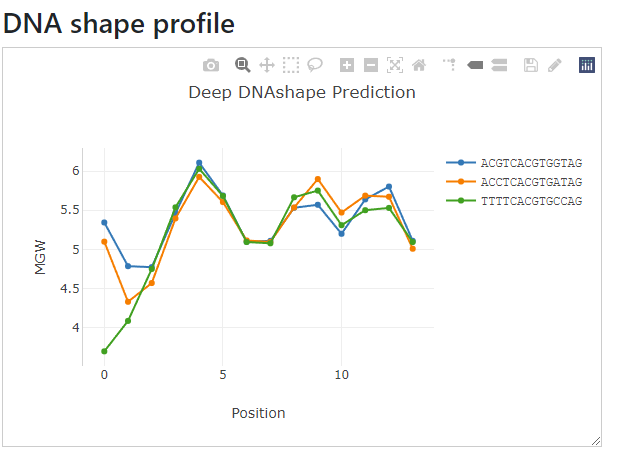
Now let's try an example with multiple sequences. In the left text input box, input more sequences (each sequence per line).
For example:
ACGTCACGTGGTAG
ACCTCACGTGATAG
TTTTCACGTGCCAG
The right side will automatically have the DNA shape profile drawn with the same order of the input.
Now, we can further edit the sequence by changing any position to another character from A, C, G, T and N, or add more sequences to compare. The right panel will reflect the change simultaneously.
File Upload and Download
Now let's try upload a text file containing a lot of DNA sequences. Create a .txt file with the following content:
ACGTCACGTGGTAG
ACCTCACGTGATAG
TTTTCACGTGCCAG
Now click 'upload' button and select the .txt file you just created. Hit the button 'Upload File & Download Predictions'.
After a couple seconds, your browser should start downloading the selected DNA shape feature predicted for the sequences in your .txt. You can now work on those DNA shape feature offline to suit your own needs.
Input Data Format
Sequence Input
Only these characters (case sensetive) are allowed: A C G T N
Input any other characters will show a warning sign and the figure will not be updated.
File Input
If you want the server to directly predict a large file containing many sequences without showing you any figures, you can upload the .txt file. Alternatively, you can upload .fa or .fasta file to the server. The server will return a downloadble link in the data format but with .txt extension.
The .txt file should contain DNA sequences with only the characters as shown above (ACGTN). Each line should contain one sequence. The maximum number of lines is 1,000,000. Files longer than 1,000,000 lines should use Deep DNAshape python package instead (Deep DNAshape).
Advanced settings
Layer Selection
Users can select the shape layer values if they want to evaluate shorter or longer flanking influence. The shape layer number means how many base pairs does the model consider in the prediction on both of the 5' and 3' ends. For example, layer 4 means 9-mer query table for intra-base-pair features and groove features, and 10-mer query table for inter-base-pair features.
Shape Features
There are a lot of shape features this web server can predict. These shape features may sound complicated. For detailed understanding of these DNA shape features, refer to our paper.
Shape Fluctuations
DNA shape fluctuations can also be predicted using this webserver. Simply select the checkbox "For shape fluctuation values, check this box." then the figure will be updated to include the fluctuation values as error bars.
DNA shape fluctuation feature is not available for EP.
Visualization and Downloading Data
Plot style change
The regular plot is a collection of line plots for multiple sequence. If you are analyzing a lot of sequences, line plots may not be a best option. Therefore, the plot style can be changed to boxplot by one-click a button 'Toggle plot type' on the top action bar.
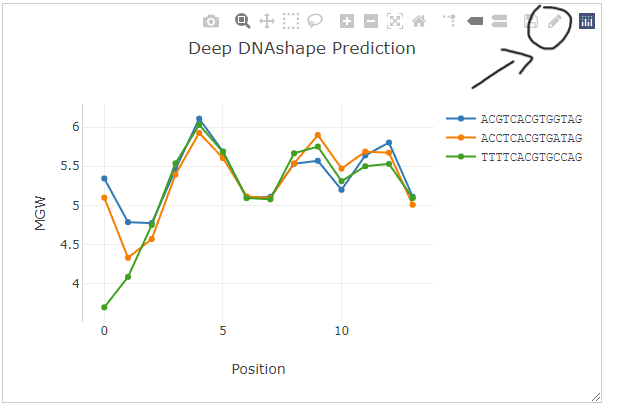
After click the button 'Toggle plot type', the plot will be changed to the following boxplot style:

Plot Customization
Plot can be resized using the right bottom corner adjuster.
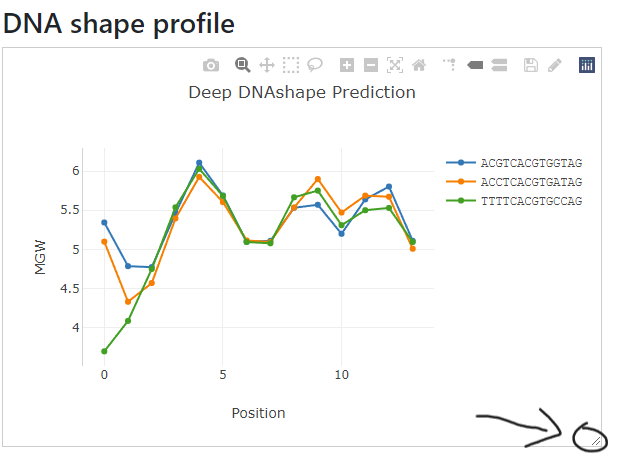
Many components in the figure are adjustable. For example, the title, x-axis and y-axis labels are editable through double click and draggable.
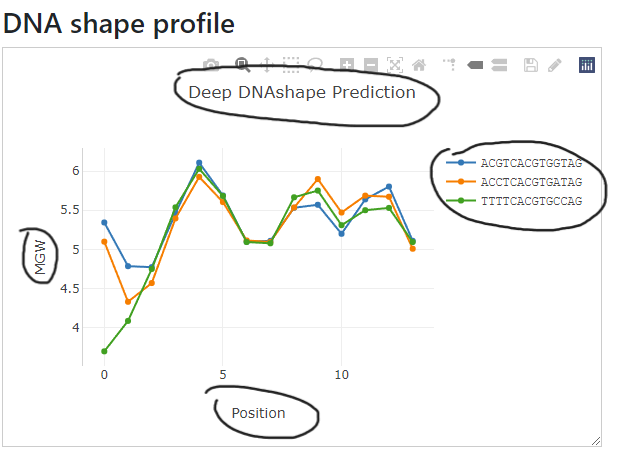
Color coding explanation for multiple sequences.
Downloading Results
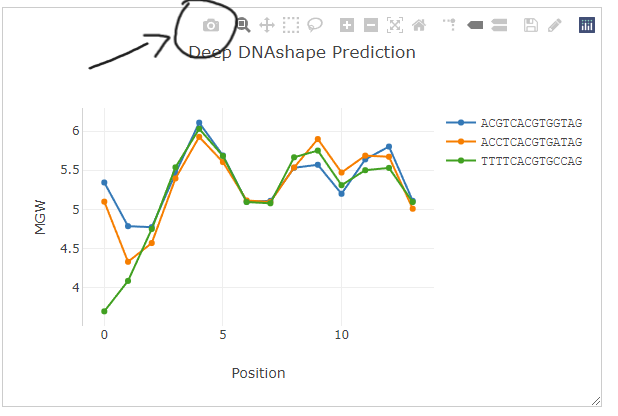
If you'd like to save the generated figure, simply click the first button on the top action bar "Download plot as a png".
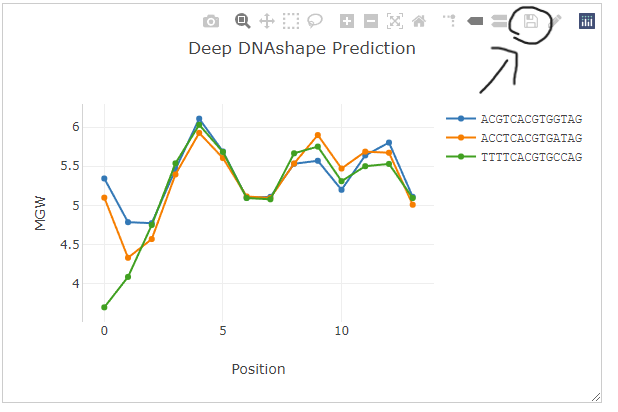
If you'd like to download all the predicted values and analyze them offline, simply click the button on the right side "Download Data". The server will return a .csv file for all the plotted data in the figure as the order of your input sequences.
About
User Privacy
User privacy is one of the top priority for our webserver. We guarantee that no IP addresses are tracked, and no cookies are stored. Uploaded files will be renamed randomly on the webserver and deleted immediately after predictions are made.
Development Team
Rohs Lab @ USC
Version History
v0.1. Release.